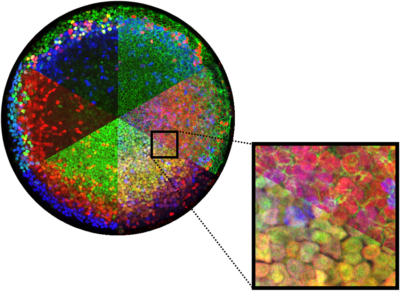
Iterative Immunofluorescence
The number of antibodies stains that can be performed on a single sample is often limiting the questions we can ask about how protein levels are related in space at the single cell level. To address this we optimized the 4i iterative immunofluorescence protocol for 2D human gastruloids and established a computational pipeline to process the data.
Find our protocol and processing pipeline for iterative IF here.
Tissue Cartography with ImSAnE (Image Surface Analysis Environment)
Advances in microscopy have enabled the study of multiscale dynamic problems such as cell tracking across entire embryos. However, increased data size makes storage, processing and transfer difficult and expensive. The data problem is most severe for specimens with curved surfaces that do not align with the microscope; in such samples, the desired information occupies only a small fraction of the region of interest (ROI) that must be recorded. Laminar data also pose conceptual problems; even when it is computationally feasible to process data in three dimensions, such as in large scale nuclear tracking, interpretation in orthographic projections or cross sections is challenging.
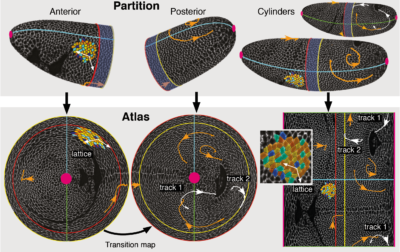
For layered samples, these problems can be solved by considering a surface of interest (SOI) instead of an ROI and move into the tissue frame by organizing the data in terms of surface coordinates. This reduces the data from 3D to 2D and is analogous to making maps of Earth that show geographic data on a flat grid of longitude and latitude.
With Sebastian Streichan, I developed a general framework for tissue cartography using differential geometry (the mathematics of curved surfaces), and a practical tool that implements it for compression and analysis of arbitrarily shaped SOIs. Our open source MATLAB toolbox ImSAnE (Image Surface Analysis Environment) maps Surfaces Of Interest to the plane, stores geometric metadata required for faithful measurements and builds an atlas containing multiple overlapping maps to create a global picture.
By now the software has been tested on a wide range of data, including:
- In toto analysis of Drosophila gastrulation
- Tracking cells on a beating zebrafish heart
- Simplified visualization of zebrafish epiboly
- Segmentation of epithelial organoids such as MDCK acini
- Nuclear segmentation in E6.5 mouse endoderm after mapping to a disc
Read about it in the Nature Methods paper. An updated version of the code can be found at github.com/idse/imsane.
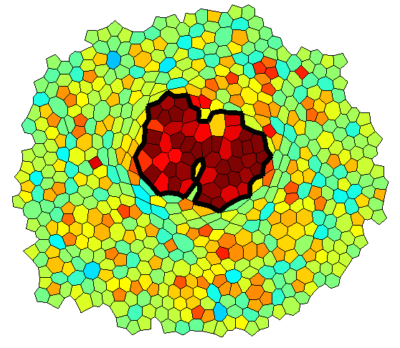
Vertex Model Simulations
Epithelial monolayers can be approximated by two-dimensional polygonal tilings that can be described by the vertex coordinates. The mechanics of the tissue can then be modeled by assigning tensions to interfaces and pressures to cells, possibly based on a model relating these properties to levels of cytoskeletal proteins. This framework includes cell rearrangement in a straightforward way through T1 transitions and simplifies cell signaling in a natural way to involve only cells and cell interfaces. I developed a simulation framework that we applied to simulating overgrowing clones in an epithelial monolayer with a mechanical feedback on myosin levels.
Read the paper that includes the simulations or download the code at github.com/idse/mechanicalFeedback.